Yeast Heat Shock Transformation on a Flexible Reconfigurable Automation Cart (RAC) System

Introduction
Authors: Pooyan Tirandazi, Ph.D., Brian Carvalho, Paulina Kanigowska Ph.D., Alexandra (Ali) Rudolph Ph.D., Alicja Ratajczak, Tom Biernacki
Baker’s yeast, Saccharomyces cerevisiae, is a workhorse molecular and synthetic biology organism serving as a fundamental research eukaryotic cell model, as well as a “chassis” for large in vivo DNA fragment construction. Yeast DNA transformation is therefore a key enabling technique used in both small-scale gene of interest expression studies, and large-scale whole-genome functional genomics studies and construction efforts, leveraging tools such as CRISPR-Cas genome editing and Transformation-associated Recombination (TAR).
Yeast DNA transformation via a heat shock is a commonly-used, cost-effective and efficient method [1]. However, despite its benefits, the overall process is complex and labor-intensive, with steps that have varying levels of automation feasibility. The complete process, even before considering the crucial downstream steps of colony picking, cell culturing, and DNA sequence verification, takes approximately 3-4 days and becomes particularly tedious when higher processing throughputs are attempted manually (10+ yeast DNA transformations / day). Additionally, it includes steps that are inherently challenging to reliably automate, notably the solid media cell culture “plating” step - an essential step that allows isolation of monoclonal cell populations, i.e. colonies, with the desired genome modifications or plasmid DNA payloads.
Recognizing these challenges, this technical note shows how Ginkgo’s Reconfigurable Automation Cart (RAC) platform can reliably automate most steps of the yeast DNA transformation process. First, we demonstrate how the core yeast heat shock DNA transformation workflow—starting from reaction setup, with manually prepared competent cells, all the way to solid media plating, employing a high-throughput “drop plating” technique—can be automated on a RAC system. We then show how the flexible architecture of RACs allows seamless system expansion to unlock the upstream competent cell preparation resulting in additional walkaway time and flexibility for scientists. Overall, the end-to-end automated process demonstrates a good balance between quality and throughput, enables more than 5 hours of walkaway time for scientists and, most importantly, offers unparalleled flexibility for scientists to schedule and run their experiments at an arbitrary time during the day/night and weekday/weekend.
Workflow
Core Transformation (Heat Shock and Solid Media Plating)
The DNA transformation process was carried out using a S. cerevisiae yeast strain from the CEN.PK family and utilized the LiAc/ssDNA/PEG technique. In this method, cells harvested during their active mid-log growth phase are rendered chemically competent when mixed with a “master mix” harboring salts (e.g. lithium acetate (LiAc) to neutralize negative charge on the DNA phosphate backbone), crowding agents (e.g. polyethylene glycol (PEG)), and denatured carrier DNA (increasing target DNA uptake), and are subjected to a “heat shock” in the presence of target DNA (linear and/or circular plasmid DNA, often designed to confer selection in addition to the intended edit) [2]. The cells are then recovered via relatively short cell culturing in liquid media over multiple hours (without any antibiotic or auxotrophic selection) and are subsequently outgrown via longer solid or liquid media cell culturing over multiple days (with antibiotic or auxotrophic selection), resulting in a subset of cells that contain the target DNA to grow on selective growth media.
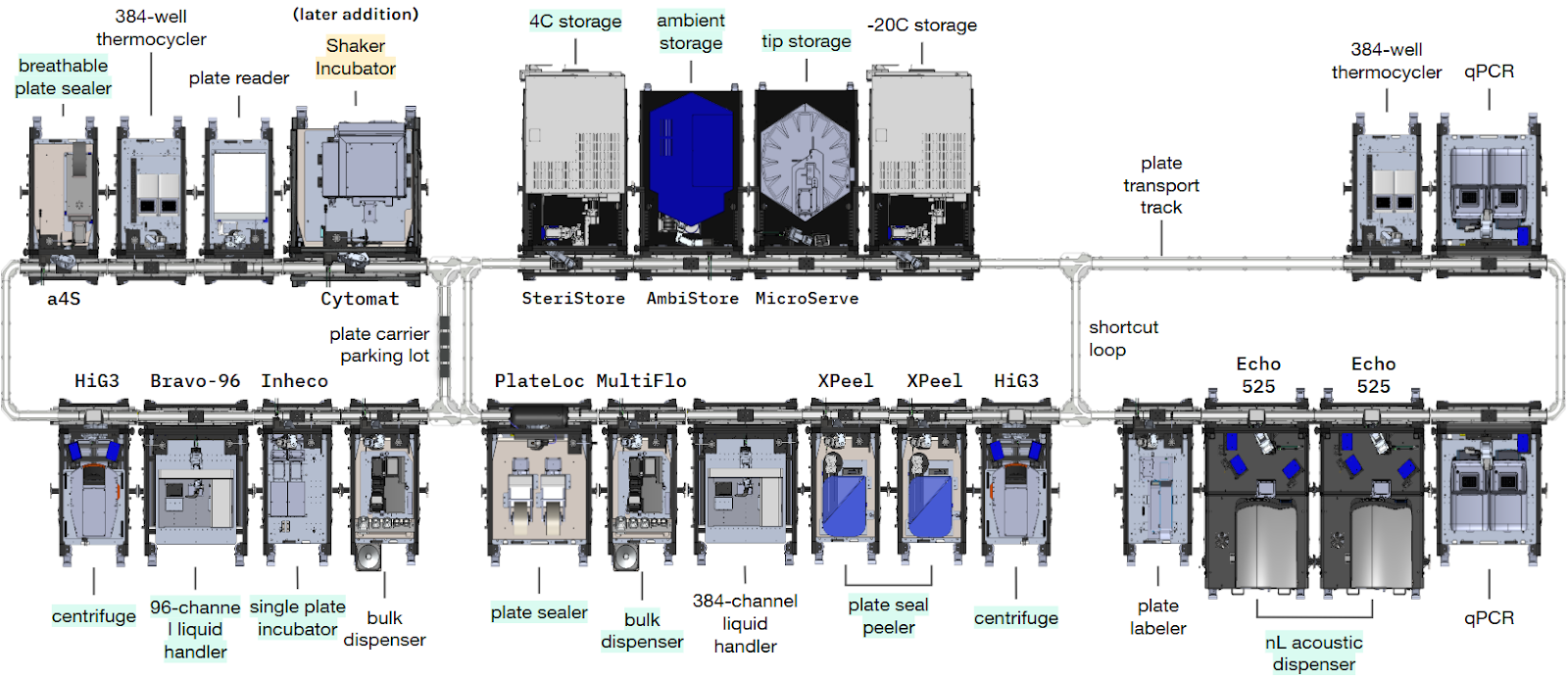
The end-to-end yeast DNA transformation workflow was divided into multiple steps as laid out in Fig 1. Each step was written as a separate protocol in Ginkgo’s Catalyst software, and was run consecutively on the RAC system at Ginkgo (Fig 2). Initial capabilities of the RAC system supported execution of most steps of the transformation workflow, except the upstream competent cell preparation and downstream solid media (agar) cell outgrowth and colony picking steps. A later expansion of the RAC system added a humidity-controlled shaking incubation capability, via a simple and quick RAC addition, which facilitated onboarding of the competent cell preparation step of the workflow. Similarly, other steps of the workflow, such as the colony picking, while out of scope for this technical note, can be onboarded with further expansion of the RAC system.
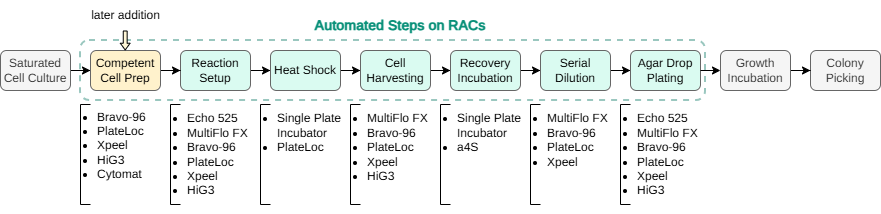
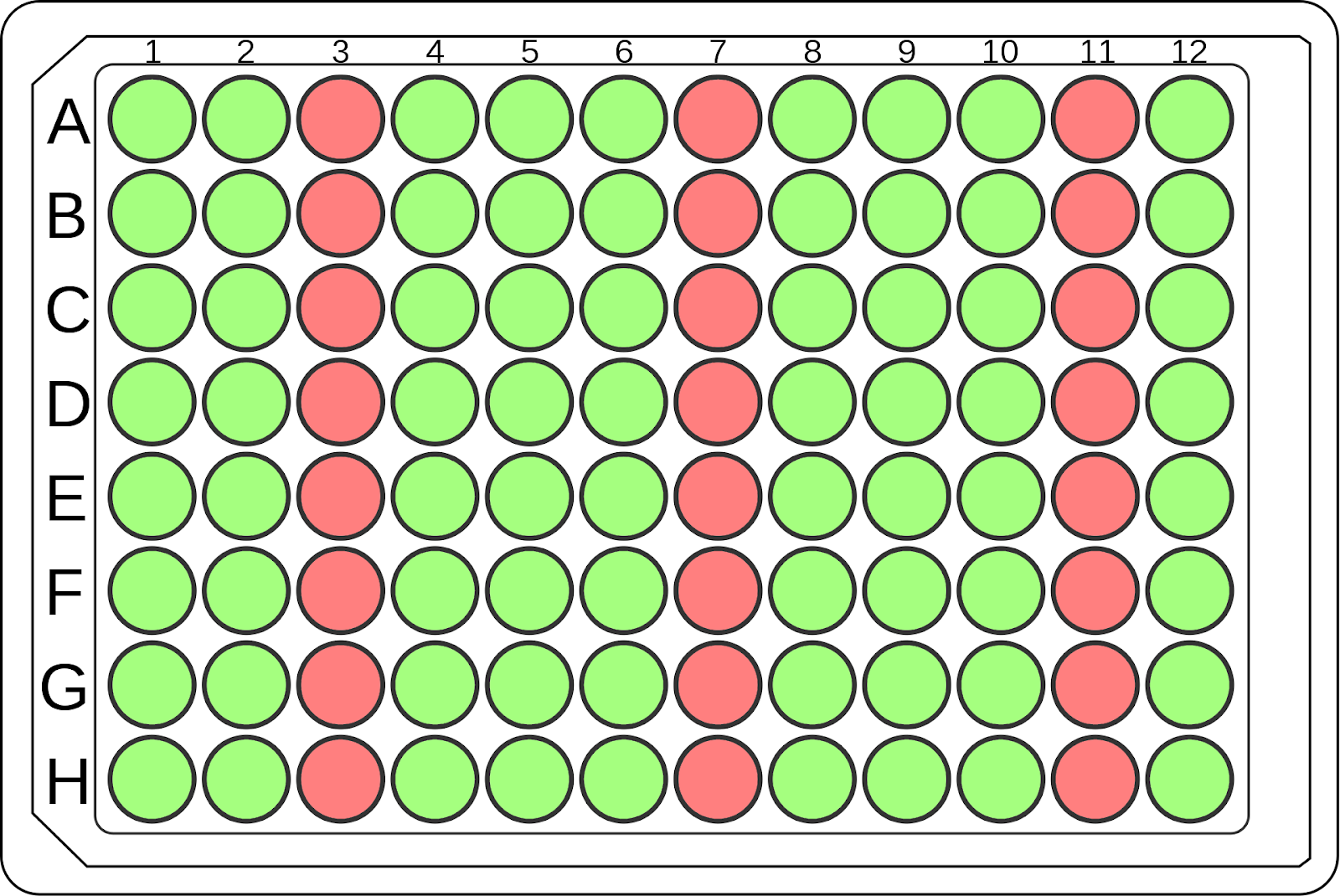
The transformation process parameters were adapted for a 96-well microplate format [3]. To evaluate the possibility of cross-contamination, the following 96-well plate layout was designed, consisting of three columns (a total of 24 wells) for negative controls that did not receive any plasmid DNA and nine columns of positive controls, representing a total of 72 technical replicates (Fig 3).
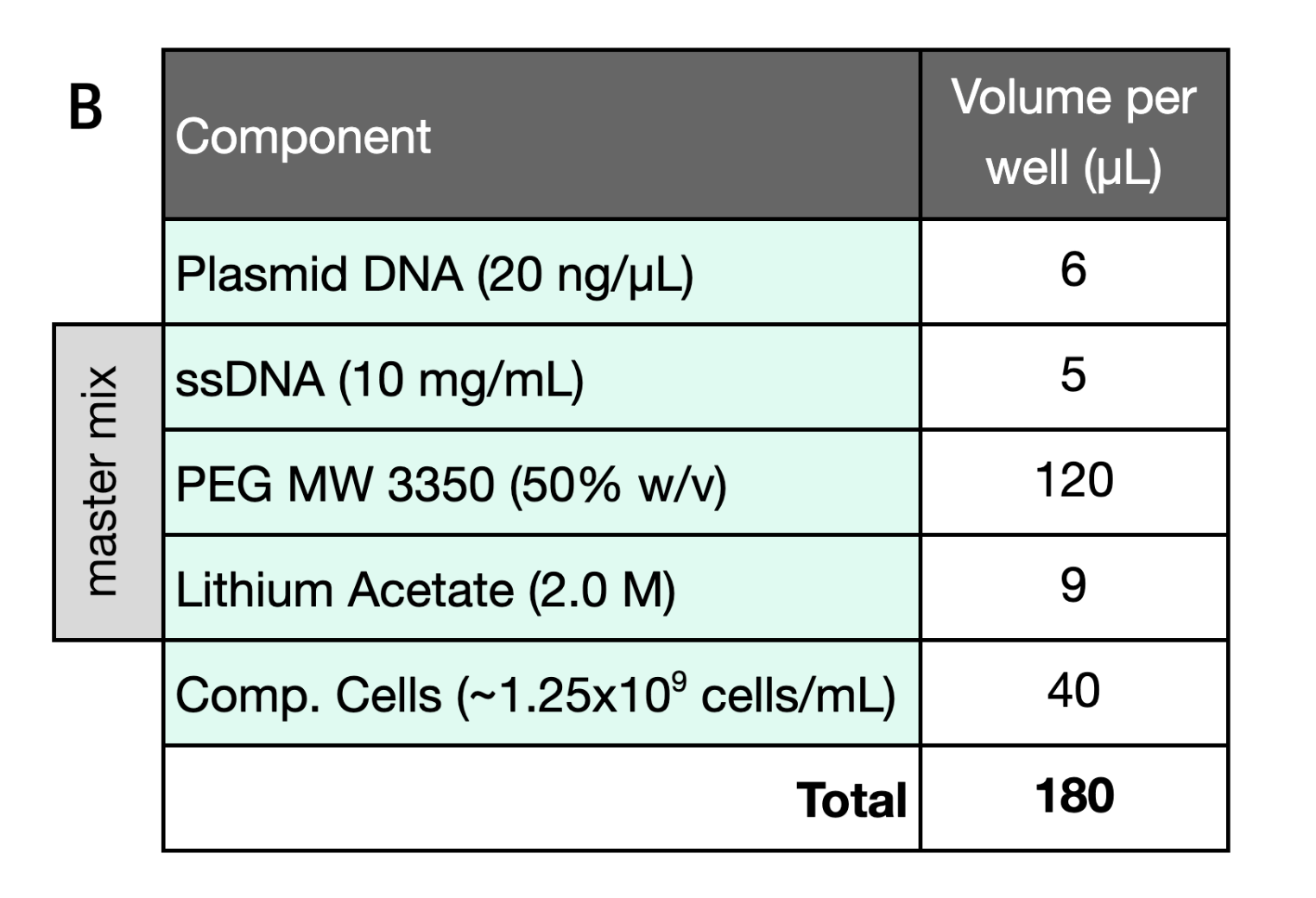
The yeast DNA transformation reaction was set up on the RAC system by combining plasmid DNA, competent cells, and the “master mix” in the “transformation plate” in the following order:
- Plasmid DNA was acoustically dispensed into the positive control plate wells (Echo 525, Beckman Coulter)
- Competent cells (previously prepared) were bulk dispensed into all plate wells (MultiFlo FX, Agilent)
- The master mix was pipette stamped and mixed into the entire plate via a 96-channel liquid handler (Bravo-96, Agilent)
After the reaction components were mixed, the sealed transformation plate was incubated in an incubator shaker (Single Plate Incubator, Inheco) on the RAC system to undergo heat shock in two stages (30 C for 30 min, followed by 42 C for 45 min). Following the heat shock step, the transformation reaction mixture was removed and the cells were washed by performing three cycles of centrifugation to pellet the cells (HiG3, BioNex), supernatant removal (Bravo-96, Agilent), bulk dispensing of a media solution (YEP+glucose (0.01% w/v)) (MultiFlo Fx, Agilent), and orbital shaking (BioShake, QInstruments) to resuspend the cells. The pelleted and washed cells were finally resuspended in the same non-selective liquid media (YEP+glucose (0.01% w/v)) that was used for washing and incubated in an incubator shaker (Single Plate Incubator, Inheco) on the RAC system for 16 hr at 30 C and 1,000 rpm, with a breathable seal (a4S, Azenta), to allow cell recovery and expression of the antibiotic resistance gene overnight.
The next day, the cells were serially diluted (10x and 100x dilution factors) using a 96-channel tip-based liquid handler (Bravo-96, Agilent) by sequentially mixing and stamping the cell suspensions into new dilution plates. The final step of the core transformation workflow consisted of solid media (agar) plating.
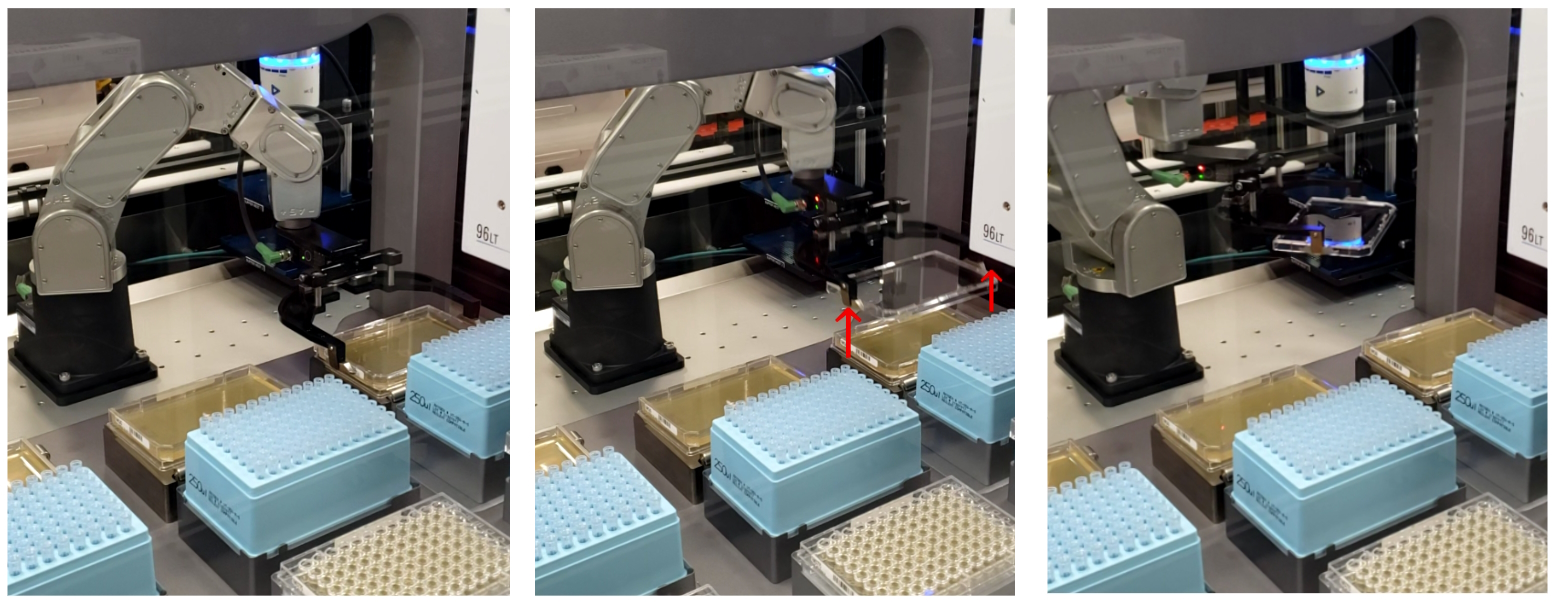
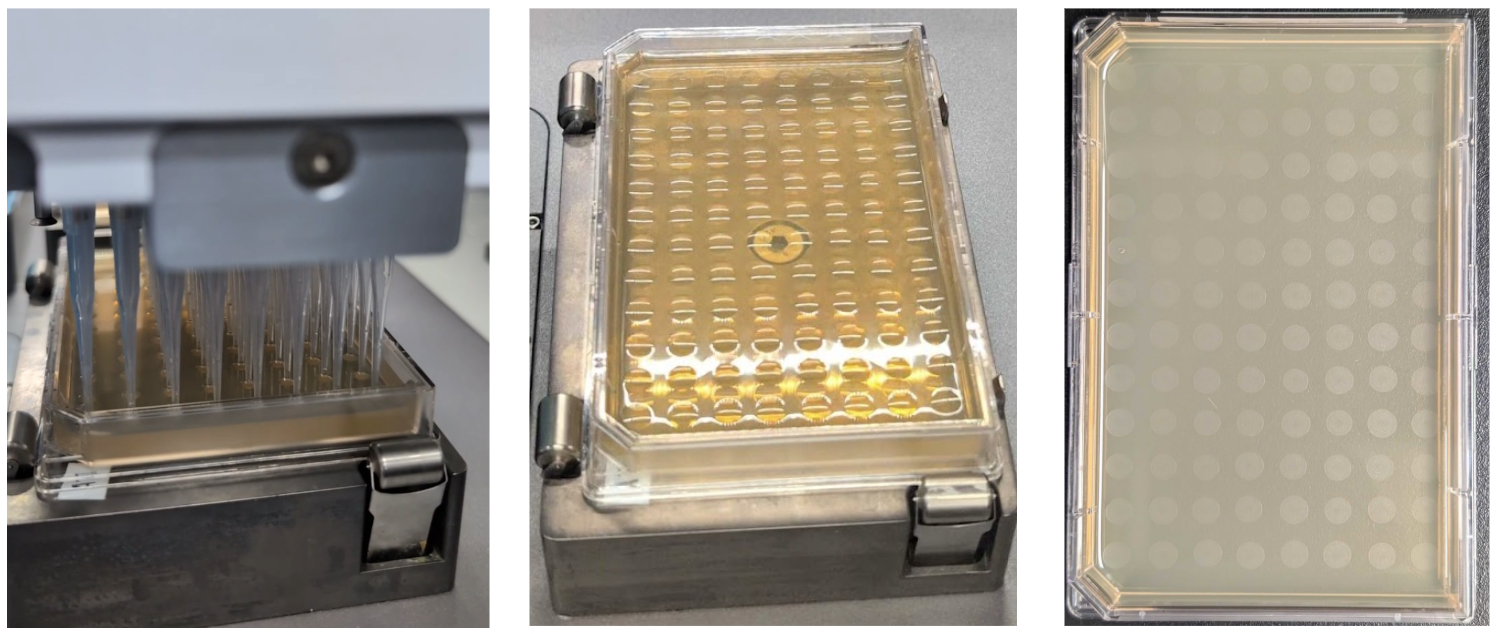
Multiple approaches exist for cell culture plating, mostly geared towards lower throughput applications. Some notable examples include, tip-based spiral plating onto Petri dishes (PetriPlater, SciRobotics), pin tool-based streak plating onto QTrays and OmniTrays (QPix, Molecular Devices), and more custom, tip-based columnwise drip plating onto tilted OmniTrays. Here, we utilized a “drop plating” technique, using tip-based stamping liquid handling, to maximize the processing throughput, while still obtaining a sufficient number of colonies needed to isolate successful “transformants”. During this step, the serially diluted plates from the previous step were loaded onto the deck of the 96-channel tip-based liquid handler (Bravo-96, Agilent) along with three SBS-format OmniTray agar plates, one for each dilution, where they were automatically delidded (Meca500, Mecademic), just prior to receiving 5 µL cell culture dilution droplets onto the agar surface. Once dry (~10 min), the OmniTrays were automatically re-lidded and transferred back to the ambient storage RAC, where an operator can pick up and incubate the agar plates for the final downstream solid media outgrowth in a humidity-controlled static incubator at 30 C for 3 days.
Competent Cell Preparation
Competent cell preparation is a key, time-constrained workflow, upstream of the core transformation workflow, setting the pace for all yeast DNA transformation steps and thus limiting execution flexibility, especially in the case of manual execution. The workflow typically starts with several hours of tedious manual work at the bench to prepare mid-log growth phase cell cultures, followed by several cell culture wash steps, and harvest of the biomass. Therefore, to increase its execution flexibility, we next sought to fully automate and integrate it with the previously developed core transformation workflow, via addition of a humidity-controlled shaking incubation RAC (Cytomat, Thermo Fisher) to the existing RAC system, which only took a day of work, and did not require any major system downtime or re-design.
Preparation of competent yeast cells was set up first by loading source plates containing saturated cell culture, rich liquid growth medium, wash buffer, and a 100 mM lithium acetate solution into ambient random access storage (AmbiStore, HighRes Biosolutions). Fresh cell cultures were next inoculated from the input saturated cell cultures using a 96-channel tip-based liquid handler (Bravo-96, Agilent), and were cultivated in a humidity-controlled shaking incubator at 30 C and 1,000 rpm (Cytomat, Thermo Fisher). Once they reached their mid-log growth phase (after ~6 hr), cultivation liquid growth medium was removed and the cells were washed via two cycles of centrifugation to pellet the cells (HiG3, BioNex), supernatant removal (Bravo-96, Agilent), bulk dispensing of water as an intermediate wash buffer (MultiFlo FX, Agilent), and orbital shaking (BioShake, QInstruments) to resuspend the cells. The final wash cycle was performed using 100 mM lithium acetate as the wash buffer, where the biomass was eventually concentrated 100x by resuspending pellets in 1% of their original cell culture volume.
Results
Automation Workflow Runtime
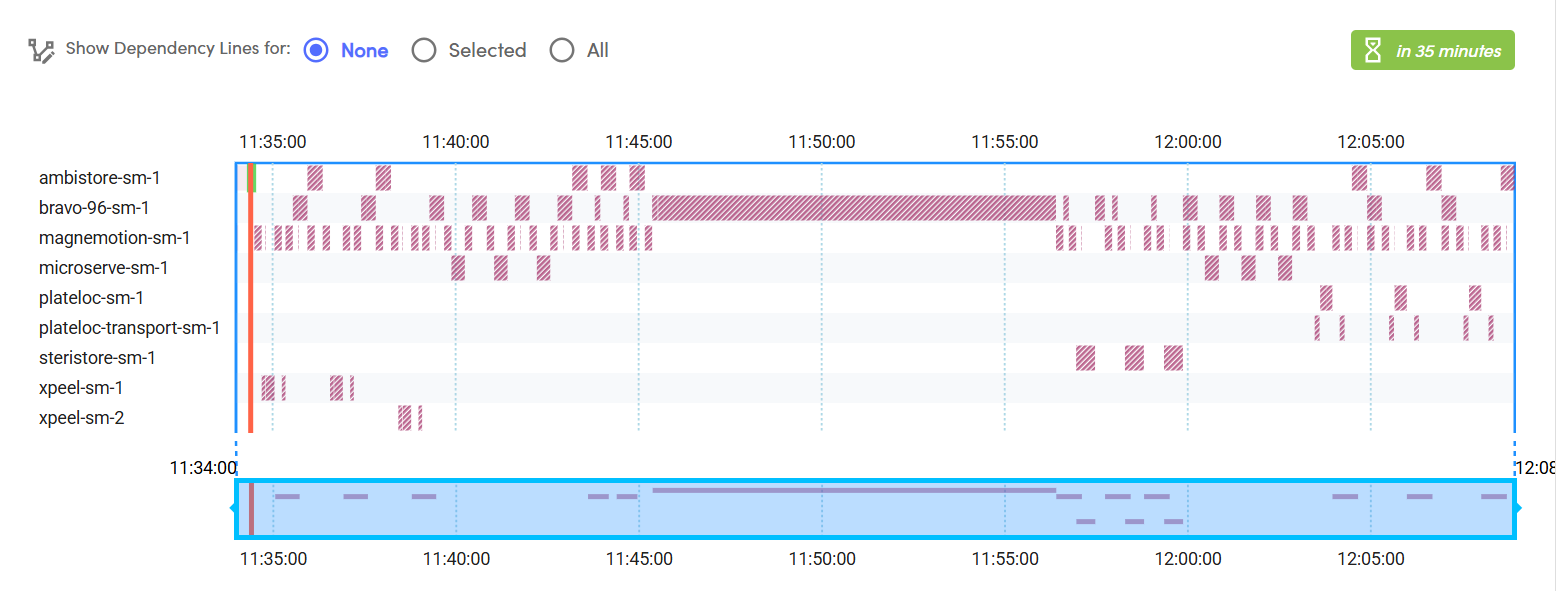
Overall, the RAC system enabled completion of 96 yeast DNA transformations - from competent cell preparation to "drop plating" - via an end-to-end, fully automated workflow. Table 1 provides a breakdown of run times for each step of the workflow on the RAC system, and an example Ginkgo Catalyst software Gantt chart schedule of the "drop plating" step is shown in Fig 4. To reduce the execution time of the workflow, the three plates for each dilution were loaded onto the liquid handler deck in batch and were processed in parallel during serial dilution and drop plating steps. Considering that the major component of the yeast heat shock DNA transformation are incubation steps (which contribute to 23 hours of non-hands-on time, split between the competent cell preparation, heat shock, and recovery step incubations), fully automating the end-to-end workflow on RACs resulted in ~5.5 hrs savings in hands-on time, during which active plate handling and liquid transfer steps took place
Drop Plating
Cell culture plating onto solid media is arguably one of the most challenging yeast DNA transformation steps to fully automate, i.e. it requires careful optimization of many different liquid transfer parameters, such as liquid dispense height (agar thickness dependent, usually > 6 mm), transfer volume (typically ~5 µL for 96-format plating), speed (typically < 5 µL/sec), and blow-out air volume (set to zero to avoid liquid splashing and cross-contamination during liquid dispense).

Ginkgo’s Catalyst software enables full parameterization of the drop plating settings along with other steps of the workflow to streamline different parameter value testing and ultimately ensure optimal performance by delivering uniform, bubble-free spots on the agar surface, not coalescing with the neighboring spots, and free from any agar surface puncture “scars”, interfering with the downstream colony imaging and picking steps (Fig 6).
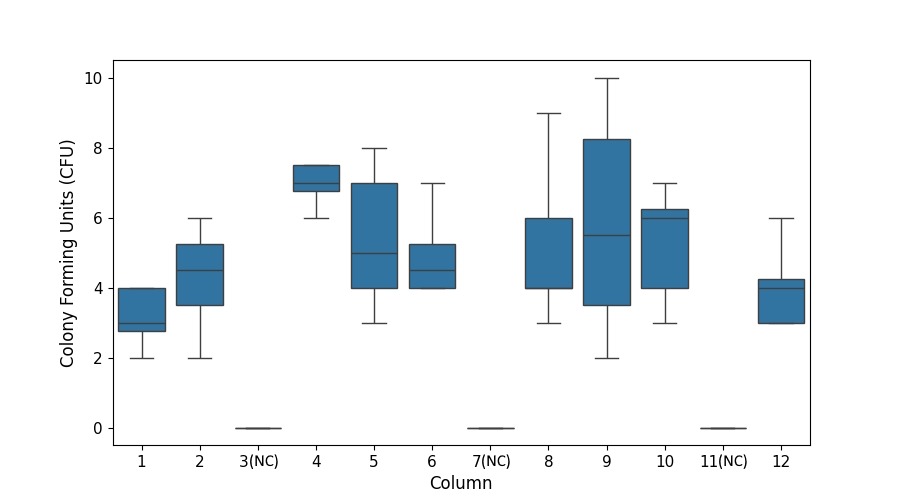
Our post-outgrowth analysis, following three days of colony outgrowth, confirmed the absence of cross-contamination in the negative control columns and no overall contamination in the positive control columns (Fig 7).
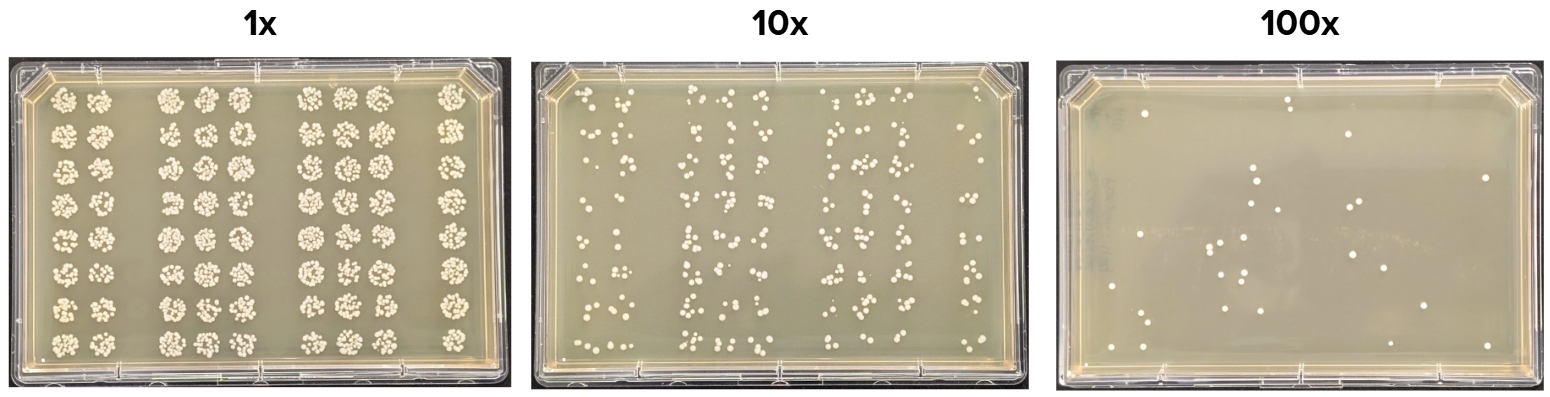
Transformation Efficiency (TE) was calculated based on the number of observed Colony Forming Units (CFUs) on the agar plate, divided by the plasmid DNA amount in the reaction (in µg), taking into account all the reaction dilutions. We were able to reliably obtain the CFU counts from the 10x dilution agar plate (Fig 8). The resulting average TE across all the technical replicates in the plate was ~ 4.24 x 104 CFU/µg.
Competent Cell Preparation
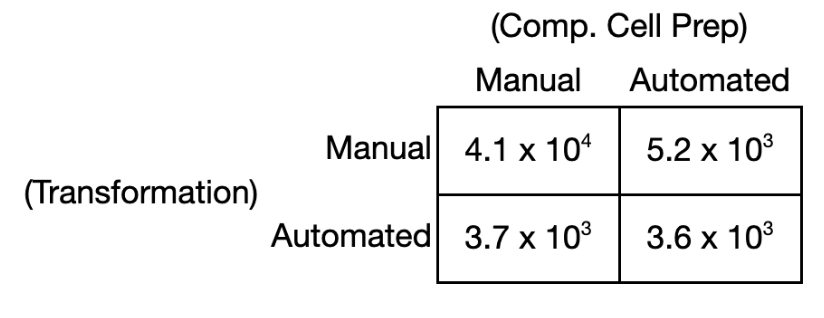
To further demonstrate that the time intensive step of competent cell preparation could be reliably automated on RACs, a separate head-to-head TE comparison was conducted between competent cells prepared on RACs vs. prepared manually (i.e. by hand at the bench). A similar comparison was made using both sources of competent cells as inputs to yeast heat shock DNA transformation performed on RACs vs. manually, ultimately creating a 2x2 grid comparison of RAC vs. manual competent cell preparation and transformation (Fig 9). The TE across the four conditions were all within a single order of magnitude. As expected, the highest TE was observed for competent cells prepared and transformed manually (TE ~4.1 x 104 CFU/µg). Despite the objectively lower TE associated with fully automated yeast DNA transformation, it is worth noting that this workflow valorized otherwise unused time by running after standard working hours and without human supervision, while still generating a sufficient number of CFUs for downstream screening.
Conclusions
This technical note demonstrates the robustness of Ginkgo's Reconfigurable Automation Cart (RAC) platform in fully automating complex, time-sensitive, and multi-step workflows. The end-to-end execution of the yeast DNA transformation workflow on RACs allowed scientists to gain over 5 hours of walkaway time in the lab. The fully automated workflow also enabled increased throughput compared to a typical manual workflow, primarily thanks to the use of 96-well format liquid handling (e.g. during cell culture “drop plating”).
While this work showcased a proof-of-concept end-to-end workflow for a single 96-well plate, larger plate batches can be scheduled on the RAC system and run in a concurrent manner by leveraging Catalyst software dynamic scheduling and interleaving capabilities. Ultimately, executing yeast DNA transformations on RACs not only boosts throughput, but also allows larger genetic engineering campaigns to be staged and launched at will and during off-peak hours (evenings or weekends), thereby valorizing scientists time and maximizing their ability to concentrate on more complex tasks.
As workflow requirements tend to evolve and scientists progressively extend the use of lab automation in their workflows, the RAC platform’s unique architecture allows seamless adaptation as needs change. Here, we demonstrated how onboarding of a humidity-controlled shaking incubation RAC allowed for a retroactive addition of the upstream competent cell preparation workflow. Future expansion of the described RAC system could further improve the described workflows or enable additional downstream steps. In particular, addition of a plate washing RAC could accelerate the multi-step cell pellet washes, during competent cell preparation, addition of an acoustic volume detection RAC could enable upstream agar surface QC, while addition of a colony picking RAC could enable downstream clone isolation and re-arraying.
References
1. R. Daniel Gietz & Robin A. Woods (2001) Genetic Transformation of Yeast, BioTechniques, 30:4,
2. Gietz RD, Schiestl RH. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc. 2007;2(1):31-4
3. Gietz RD, Schiestl RH. Microtiter plate transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc. 2007;2(1):5-8
Materials
- Plasmid DNA: Cas9 expression plasmid with truncated Hygromycin antibiotic marker, 10.2 kbp
- YEP Broth (Teknova, cat. no. Y3026)
- 40% Glucose Solution (Teknova, cat. no. G2021)
- 50% Polyethylene Glycol 3350 (w/v) (Boston Bioproducts Inc., cat. no. C-9999G)
- Lithium Acetate Dihydrate (Sigma-Aldrich, cas. no. 6108-17-4)
- Sheared Salmon Sperm DNA (10 mg/mL) (Thermo Fisher Scientific, cat. no. AM9680)
- YPD Agar with Hygromycin-250 (prepared in house) and poured in OmniTray™ Single-Well Plate with Lid (Thermo Fisher Scientific, cat. no. 242811)
- 96-well Clear Round Bottom TC-treated Microplate (Corning, cat. no. 3798)
- 96-well Clear V-Bottom 2mL Polypropylene Deep Well Plate (Corning, cat. no. 3960)
- Echo Qualified 384-Well Polypropylene Microplates (Beckman Coulter, cat. no. PP-0200)
- 96-Well Tips, 250µL, Clear, Non-filtered (Axygen, cat. no. VT-250-R-S)
- 2L Baffled Erlenmeyer Flask (Thermo Fisher Scientific, cat. no. 4116-2000 )
- 125mL Baffled Erlenmeyer Flask (Thermo Fisher Scientific, cat. no. 4116-0125)
{{data}}
Subscribe to receive future tech notes
- 100% Hands-free yeast transformation, including plating on solid media
- +5hrs Of walkaway time for scientists
- <1 day RAC system expansion to enable additional competent cell prep
Real Labs. Real Impact.
Learn more about our technology notes.


